Transvection Is Common Throughout the Drosophila Genome
authors: David J Mellert, James W Truman
doi: 10.1534/genetics.112.140475
CITATION
Mellert, D. J., & Truman, J. W. (2012). Transvection Is Common Throughout the Drosophila Genome. Genetics, 191(4), 1129–1141. https://doi.org/10.1534/genetics.112.140475
ABSTRACT
Higher-order genome organization plays an important role in transcriptional regulation. In Drosophila, somatic pairing of homologous chromosomes can lead to transvection, by which the regulatory region of a gene can influence transcription in trans. We observe transvection between transgenes inserted at commonly used phiC31 integration sites in the Drosophila genome. When two transgenes that carry endogenous regulatory elements driving the expression of either LexA or GAL4 are inserted at the same integration site and paired, the enhancer of one transgene can drive or repress expression of the paired transgene. These transvection effects depend on compatibility between regulatory elements and are often restricted to a subset of cell types within a given expression pattern. We further show that activated UAS transgenes can also drive transcription in trans. We discuss the implication of these findings for (1) understanding the molecular mechanisms that underlie transvection and (2) the design of experiments that utilize site-specific integration.
fleeting notes
consider transvection when designing fly crosses
one of the joys of using flies is the extensive and specific genetic access we have. however, there are some caveats to the tools we use. attP sites were introduced into the genome through P-element transformations. these attP sites enable site directed insertion of transgenes to known locations.
important :
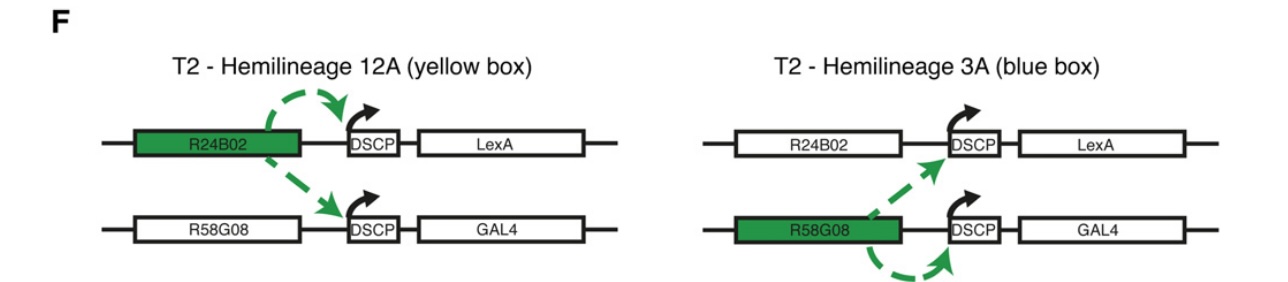
when using intersectional approaches with two different binary expression systems (Gal4 & LexA) together in the fly, transvection requires deliberate consideration! If your LexA and Gal4 drivers are integrated at the same attP site, the promotors may be used to drive trans expression of the opposite transcription factor (x-Gal4 may become x-LexA and y-LexA may become y-Gal4)
fig2F
schematic depicting transvection between transgenes when at the same attP site
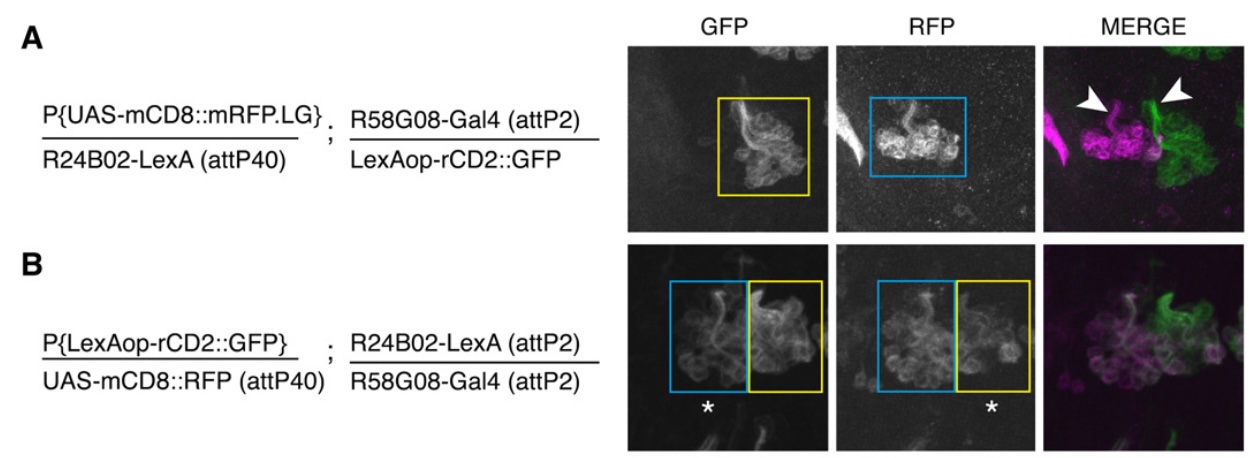
fig2a,b
a) is the control. only one group of neurons is labeled by each reporter (GFP vs RFP) because different attp sites were used for the drivers
b) the result of transvection. you get expression of both neuron groups by each reporter