when analyzing connectomes like FAFB, FANC, MANC, and BANC there are often specific neurons that you find and hypothesize that they are important for some behavior. However, testing this experimentally requires having access to those neurons. luckily there are a number of ways to find genetic driver lines (like gal4 lines or split gal4 lines) that label your neurons of interest! below are a few thoughts about how I go about finding driver lines.
this process has become a lot easier recently. gone are the days of manually sorting through thousands of light microscopy images with GFP labeled cells. there are tools that do this searching for you which speeds things up tremendously.
The best way to jump between electron microscopy and light microscopy is neuronbridge. the huge light microscopy datasets collected by the flylight team at janelia have been registered in 3 dimensions and can be compared to multiple public connectome datasets (right now this works with the janelia hemibrain dataset, flywire brain, manc, and soon to be the banc dataset). This tool generates a colormap on the neuron based on the Z depth and then finds the closest matching light microscopy images based on those pixels. It generally works pretty well, although many have found driver lines still with the old fashioned way of sifting through images by hand.
On neuron bridge you can search for your neuron based on the ID from the dataset you are using (fafb/manc). then two results pop up. select the button that says “Color Depth Search Results” for the option that matches the neuron ID and dataset that you are starting from.
see this example for DNg02 (10240 in MANC):
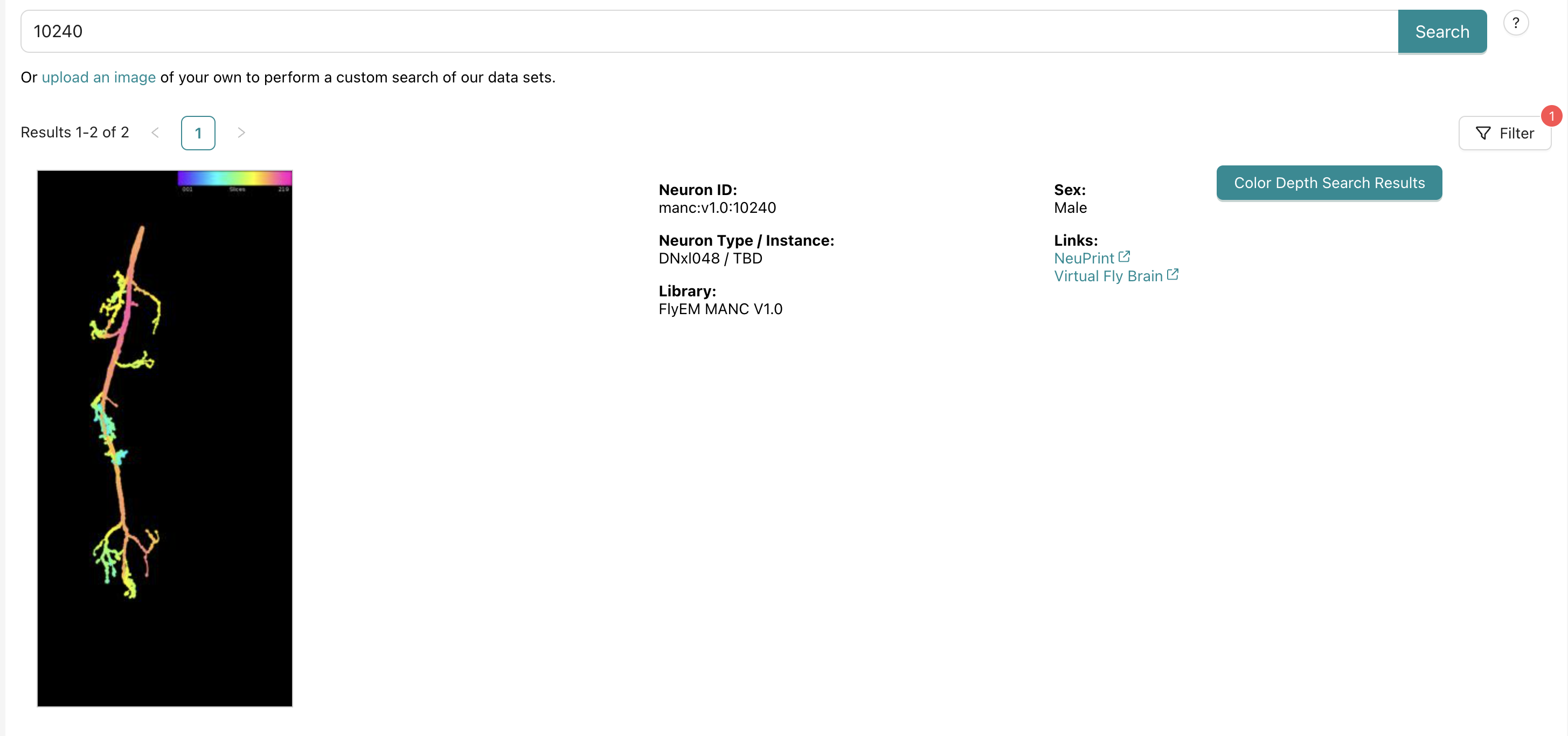
this will return a bunch of light microscopy images that have a score. the score comes from matched pixels from the color depth search.
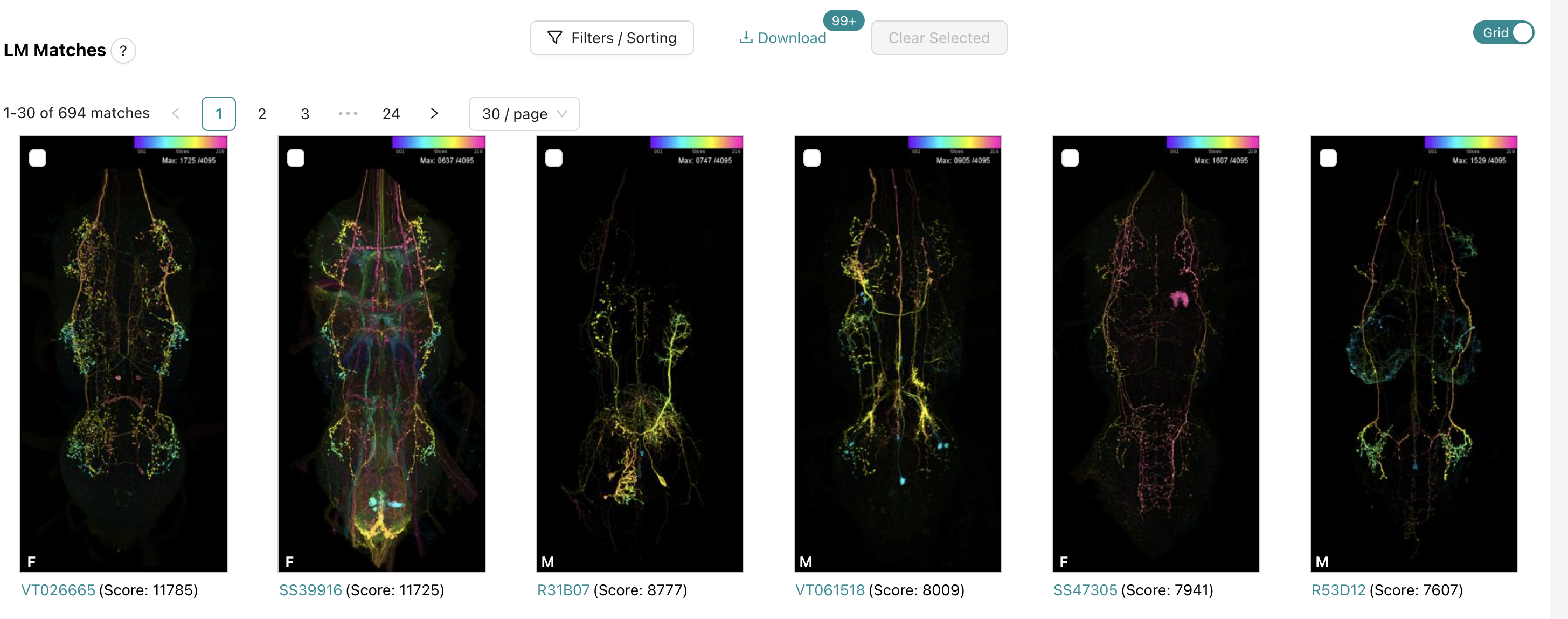
already, the 1st and 2nd results looks pretty. good. The second result is a split gal4 line, so there might be pretty sparse labeling of neurons which can be really helpful. clicking on this second result lets us examine it closer
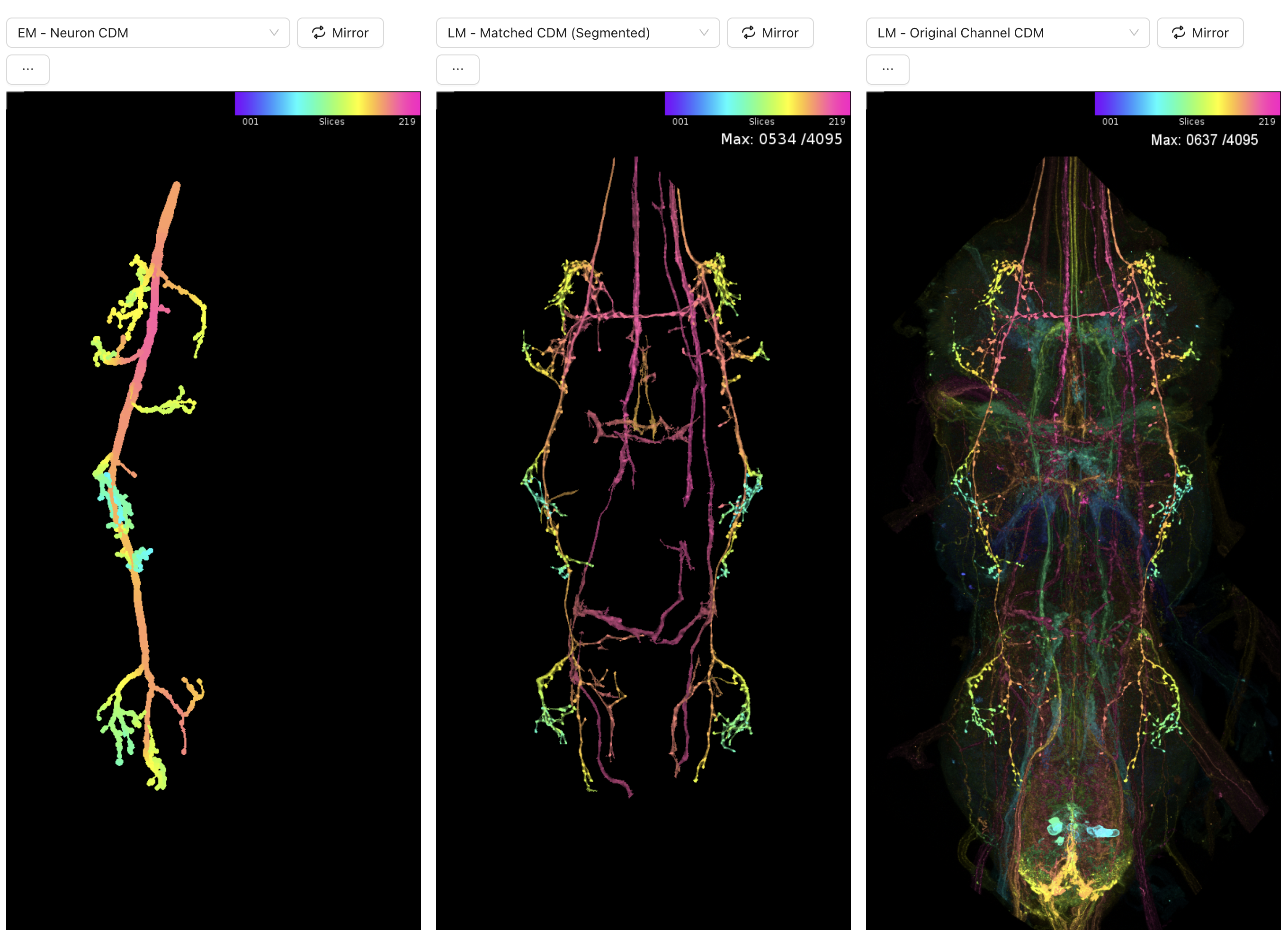
the neuron on the left is the EM neuron from the manc dataset. the middle and right images are from the light microscopy images. this looks like a really really good match morphologically! you now have a driver line that labels your neuron of interest and you can see what the name of it is at the top:
alternatives
what do you do if you are not working with one of the datasets that neuron bridge is ready to accept? The best way to find a driver line for your neuron is to first find your neuron in one of those datasets that neuron bridge accepts. This can be simple or complicated depending on the neuron. One way to start is to use the cell type information to begin narrowing down the possible cells in the other connectome dataset.
another tool you can try to use is called braincircuits which has some tools built in to find flylight matches to your EM neurons. I havent used this tool much personally but I think it could be a great option!